BULLETIN 1
From science to action: Advancing the Global Biodiversity Framework at SBSTTA-27

CGIAR October 22 2025
By Murillo Adelaida Leon
As the world’s biodiversity losses continue, the need for evidence-based action and cross-sector collaboration has never been more urgent. Biodiversity is fundamental to building climate-resilient, sustainable food, land, and water systems, and to maintain the ecological functions that sustain human well-being.
This week, the 27th meeting of the Subsidiary Body on Scientific, Technical and Technological Advice (SBSTTA-27) is taking place in Panama City, as one of the key preparatory milestones on the road to COP17. During the meeting, countries are taking stock of progress in implementing the Kunming–Montreal Global Biodiversity Framework (KMGBF) and identify pathways to accelerate action.
SBSTTA-27, together with the new Subsidiary Body on Article 8(j) (focused on Indigenous Peoples and local communities), represents the most comprehensive review process in the Convention on Biological Diversity (CBD) since COP15.
Bringing together scientists, policymakers, and stakeholders from across the globe, SBSTTA-27 aims to deliver science-based recommendations to guide GBF implementation and inform decisions for COP17 in Yerevan, Armenia, from 19 to 30 October 2026.
For the CGIAR Multifunctional Landscapes (MFL) Science Program, this meeting represents both a scientific and strategic opportunity to strengthen its role and contributions toward integrated, landscape-based approaches on the road to COP17. It also provides a valuable space to actively listen to the needs and knowledge gaps expressed by Parties, ensuring that our research remains aligned with global priorities.
See https://www.cgiar.org/news-events/news/from-science-to-action-advancing-the-global-biodiversity-framework-at-sbstta-27/
As the world’s biodiversity losses continue, the need for evidence-based action and cross-sector collaboration has never been more urgent. Biodiversity is fundamental to building climate-resilient, sustainable food, land, and water systems, and to maintain the ecological functions that sustain human well-being.
This week, the 27th meeting of the Subsidiary Body on Scientific, Technical and Technological Advice (SBSTTA-27) is taking place in Panama City, as one of the key preparatory milestones on the road to COP17. During the meeting, countries are taking stock of progress in implementing the Kunming–Montreal Global Biodiversity Framework (KMGBF) and identify pathways to accelerate action.
SBSTTA-27, together with the new Subsidiary Body on Article 8(j) (focused on Indigenous Peoples and local communities), represents the most comprehensive review process in the Convention on Biological Diversity (CBD) since COP15.
Bringing together scientists, policymakers, and stakeholders from across the globe, SBSTTA-27 aims to deliver science-based recommendations to guide GBF implementation and inform decisions for COP17 in Yerevan, Armenia, from 19 to 30 October 2026.
For the CGIAR Multifunctional Landscapes (MFL) Science Program, this meeting represents both a scientific and strategic opportunity to strengthen its role and contributions toward integrated, landscape-based approaches on the road to COP17. It also provides a valuable space to actively listen to the needs and knowledge gaps expressed by Parties, ensuring that our research remains aligned with global priorities.
See https://www.cgiar.org/news-events/news/from-science-to-action-advancing-the-global-biodiversity-framework-at-sbstta-27/
BULLETIN 2
Scientists Discover Pair of Genes that Strengthens Wheat’s Defense Against Powdery Mildew
Scientists Discover Pair of Genes that Strengthens Wheat’s Defense Against Powdery Mildew

Scientists have identified a pair of resistance genes in wild emmer wheat that could help protect cultivated varieties from powdery mildew, a major fungal disease affecting global wheat yields. The study emphasized that two adjacent NLR proteins, PmWR183-NLR1 and PmWR183-NLR2, must work together to confer immunity.
The team used advanced techniques, including map-based cloning, long-read genome sequencing, and CRISPR-Cas9, to uncover how PmWR183 functions. Unlike most known resistance genes, PmWR183 depends on the cooperation of two NLR proteins that physically interact to trigger the plant's immune response. Researchers found that PmWR183-NLR2 shows high diversity in its sequence, possibly reflecting a long evolutionary battle between wheat and its pathogens.
Notably, PmWR183 provides strong resistance at the adult stage of plant growth but limited protection in seedlings. Tests confirmed that this stage-dependent resistance is linked to changing expression levels of the two NLR genes as the plant matures. The researchers suggest that increasing PmWR183-NLR2 activity could extend protection earlier in development and help create wheat varieties that resist disease throughout their life cycle.
For more information, read the information from Nature.
See https://www.isaaa.org/kc/cropbiotechupdate/article/default.asp?ID=21556
SCIENTIFIC NEWS
Intra species dissection of Phytophthora capsici resistance in black pepper
Yupeng Hao, Rui Fan, Yongyan Zhao, Ke Nie, Luyao Wang, Ting Zhao, Zhiyuan Zhang, Xiaoyuan Tao, Hongyu Wu, Jiaying Pan, Chaoyun Hao, Xueying Guan
J Adv Res.; 2025 Aug: 74:121-136. doi: 10.1016/j.jare.2024.10.015.
Intra species dissection of Phytophthora capsici resistance in black pepper
Yupeng Hao, Rui Fan, Yongyan Zhao, Ke Nie, Luyao Wang, Ting Zhao, Zhiyuan Zhang, Xiaoyuan Tao, Hongyu Wu, Jiaying Pan, Chaoyun Hao, Xueying Guan
J Adv Res.; 2025 Aug: 74:121-136. doi: 10.1016/j.jare.2024.10.015.

ABSTRACT
Introduction: Black pepper, a financially significant tropical crop, assumes a pivotal role in global agriculture for the major source of specie flavor. Nonetheless, the growth and productivity of black pepper face severe impediments due to the destructive pathogen Phytophthora capsici, ultimately resulting in black pepper blight. The dissecting for the genetic source of pathogen resistance for black pepper is beneficial for its global production. The genetic sources include the variations on gene coding sequences, transcription capabilities and epigenetic modifications, which exerts hierarchy of influences on plant defense against pathogen. However, the understanding of genetic source of disease resistance in black pepper remains limited.
Methods: The wild species Piper flaviflorum (P. flaviflorum, Pf) is known for blight resistance, while the cultivated species P. nigrum is susceptible. To dissecting the genetic sources of pathogen resistance for black pepper, the chromatin modification on H3K4me3 and transcriptome of black pepper species were profiled for genome wide comparative studies, applied with CUT&Tag and RNA sequencing technologies.
Results: The intraspecies difference between P. flaviflorum and P. nigrum on gene body region led to coding variations on 5137 genes, including 359 gene with biotic stress responses and regulation. P. flaviflorum exhibited a more comprehensive resistance response to Phytophthora capsici in terms of transcriptome features. The pathogen responsive transcribing was significant associated with histone modification mark of H3K4me3 in black pepper. The collective data on variations of sequence, transcription activity and chromatin structure lead to an exclusive jasmonic acid-responsive pathway for disease resistance in P. flaviflorum was revealed. This research provides a comprehensive frame work to identify the fine genetic source for pathogen resistance from wild species of black pepper.
See https://pubmed.ncbi.nlm.nih.gov/39442874/
Methods: The wild species Piper flaviflorum (P. flaviflorum, Pf) is known for blight resistance, while the cultivated species P. nigrum is susceptible. To dissecting the genetic sources of pathogen resistance for black pepper, the chromatin modification on H3K4me3 and transcriptome of black pepper species were profiled for genome wide comparative studies, applied with CUT&Tag and RNA sequencing technologies.
Results: The intraspecies difference between P. flaviflorum and P. nigrum on gene body region led to coding variations on 5137 genes, including 359 gene with biotic stress responses and regulation. P. flaviflorum exhibited a more comprehensive resistance response to Phytophthora capsici in terms of transcriptome features. The pathogen responsive transcribing was significant associated with histone modification mark of H3K4me3 in black pepper. The collective data on variations of sequence, transcription activity and chromatin structure lead to an exclusive jasmonic acid-responsive pathway for disease resistance in P. flaviflorum was revealed. This research provides a comprehensive frame work to identify the fine genetic source for pathogen resistance from wild species of black pepper.
See https://pubmed.ncbi.nlm.nih.gov/39442874/
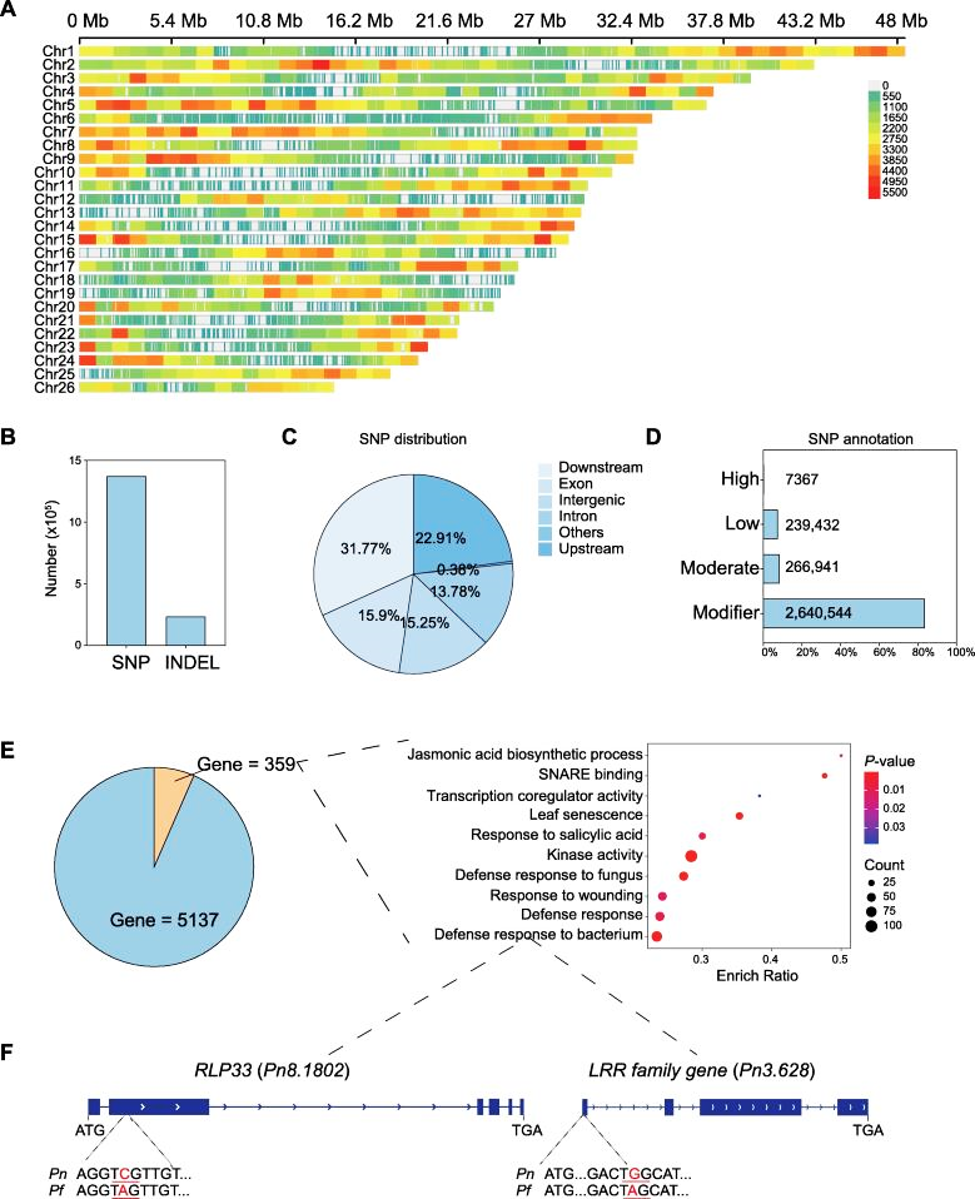
Figure:
The genomic differences on transcribed region in disease resistance in Piper flaviflorum.
The genomic differences on transcribed region in disease resistance in Piper flaviflorum.
A: The distribution of SNPs on chromosomes of Pf. The heatmap shows the density of SNPs. B: The number of SNPs and INDELs of Pf. C: Pie chart of the SNPs distribution by genes’ region. Different color blocks represent different regions. D: SNPs annotation of the effects by impact. HIGH: The variant is assumed to have high (disruptive) impact in the protein, probably causing protein truncation, loss of function or triggering nonsense mediated decay. Moderate:A non-disruptive variant that might change protein effectiveness. Low: Assumed to be mostly harmless or unlikely to change protein behavior. Modifier: Usually non-coding variants or variants affecting non-coding genes, where predictions are difficult or there is no evidence of impact. E: Pie chart and the GO terms of genes related to disease resistance in high-impact level genes. F: The SNP and gene structure of the candidate genes. Pn represents Piper nigrum and Pf represents Piper flaviflorum.
.png)











