BULLETIN 1
How rice + seaweed can transform food and sustainability efforts
How rice + seaweed can transform food and sustainability efforts
.png)
CGIAR October 20 2025
By Glenn Concepcion
In a world grappling with a rising population, climate change, and a growing need for healthier food options, scientists are turning to an unlikely pairing: rice and seaweed.
A scientific review published in Critical Reviews in Food Science and Nutrition explores the vast, untapped potential of combining rice starch with biopolymers from seaweed to create transformative products that benefit both human and planetary health.
Rice is a staple food for more than half the global population, but a significant portion—over 15% of polished rice—consists of broken grains often sold for lower-value uses like animal feed. At the same time, seaweed production has skyrocketed, growing from just half a million wet tons in 1950 to nearly 36 million tons in 2020, thanks to its exceptional nutritional profile rich in fiber, vitamins, minerals, and antioxidants.
Researchers at the International Rice Research Institute and the University of the Philippines-Diliman Marine Science Institute saw a powerful opportunity in these two resources. By extracting starch from underutilized broken rice grains and blending it with hydrocolloids—natural gelling agents like agar, alginate, and carrageenan from seaweed—they can create novel materials with remarkable properties.
“Combining rice starch with seaweed biopolymers is not just a technical novelty,” the study’s authors state, “it reflects a strategic alignment of nutritional enhancement, material functionality, and environmental sustainability”.
See https://www.cgiar.org/news-events/news/how-rice-seaweed-can-transform-food-and-sustainability-efforts
In a world grappling with a rising population, climate change, and a growing need for healthier food options, scientists are turning to an unlikely pairing: rice and seaweed.
A scientific review published in Critical Reviews in Food Science and Nutrition explores the vast, untapped potential of combining rice starch with biopolymers from seaweed to create transformative products that benefit both human and planetary health.
Rice is a staple food for more than half the global population, but a significant portion—over 15% of polished rice—consists of broken grains often sold for lower-value uses like animal feed. At the same time, seaweed production has skyrocketed, growing from just half a million wet tons in 1950 to nearly 36 million tons in 2020, thanks to its exceptional nutritional profile rich in fiber, vitamins, minerals, and antioxidants.
Researchers at the International Rice Research Institute and the University of the Philippines-Diliman Marine Science Institute saw a powerful opportunity in these two resources. By extracting starch from underutilized broken rice grains and blending it with hydrocolloids—natural gelling agents like agar, alginate, and carrageenan from seaweed—they can create novel materials with remarkable properties.
“Combining rice starch with seaweed biopolymers is not just a technical novelty,” the study’s authors state, “it reflects a strategic alignment of nutritional enhancement, material functionality, and environmental sustainability”.
See https://www.cgiar.org/news-events/news/how-rice-seaweed-can-transform-food-and-sustainability-efforts
BULLETIN 2
Media Practitioners and Biotech Experts Discuss Biotech Updates in the Philippines
Media Practitioners and Biotech Experts Discuss Biotech Updates in the Philippines
.png)
Photo Source: CropLife Philippines
Over 40 media practitioners and key stakeholders participated in the Media Forum on Biotechnology R&D and Regulatory Landscape in the Philippines, organized by Science Communicators Philippines (SciCommPH), CropLife Philippines, and the Southeast Asian Regional Center for Graduate Study and Research in Agriculture (SEARCA). Held on October 14, 2025, at the Century Park Hotel, Manila, the forum aimed to provide journalists with science-based updates, direct access to experts, and a platform for dialogue to strengthen media reporting on agri-biotech.
The plenary session focused on global and local updates about the current agri-biotech landscape. Ms. Kristine Grace Tome shared the 2024 global data of GM crop adoption, the recently approved GM crops and their traits, the GM crops in the pipeline, and gene-editing breakthroughs in plants and animals.
“I hope that as we fulfill our pact for the future, we take a hopeful perspective on biotech for our agriculture and food. Biotech is an innovation designed to contribute to food security and other pressing global challenges. Let us harness this technology and provide safeguards to ensure that the benefits of biotechnology reach those who need it most,” said Ms. Tome.
Mr. Joy Bartolome Duldulao of the Philippine Rice Research Institute (PhilRice) and Dr. Lourdes Taylo of the UPLB Institute of Plant Breeding also tackled Malusog Rice and Bt eggplant, respectively. Dr. Clarisse Gonzalvo from the University of the Philippines Los Baños (UPLB) College of Development Communication (Devcom) shared the findings of her survey on biotech farmers' perception of the 2015 biotech ban
“If the precautionary principle tells us ‘when in doubt, stop,' then social science tells us ‘when in doubt, listen'—listen to the farmers whose lives depend on these decisions,” said Dr. Gonzalvo.
A panel discussion with experts from the Bureau of Plant Industry, the Department of Agriculture Biotechnology Program Office, and PhilRice delved into policies and programs that may help catalyze biotech adoption. Dr. Winifredo Dagli, Chair of the UPLB Devcom's Department of Science Communication, moderated the discussion.
For more information, send an email to knowledgecenter@isaaa.org
See https://www.isaaa.org/kc/cropbiotechupdate/article/default.asp?ID=21552
Over 40 media practitioners and key stakeholders participated in the Media Forum on Biotechnology R&D and Regulatory Landscape in the Philippines, organized by Science Communicators Philippines (SciCommPH), CropLife Philippines, and the Southeast Asian Regional Center for Graduate Study and Research in Agriculture (SEARCA). Held on October 14, 2025, at the Century Park Hotel, Manila, the forum aimed to provide journalists with science-based updates, direct access to experts, and a platform for dialogue to strengthen media reporting on agri-biotech.
The plenary session focused on global and local updates about the current agri-biotech landscape. Ms. Kristine Grace Tome shared the 2024 global data of GM crop adoption, the recently approved GM crops and their traits, the GM crops in the pipeline, and gene-editing breakthroughs in plants and animals.
“I hope that as we fulfill our pact for the future, we take a hopeful perspective on biotech for our agriculture and food. Biotech is an innovation designed to contribute to food security and other pressing global challenges. Let us harness this technology and provide safeguards to ensure that the benefits of biotechnology reach those who need it most,” said Ms. Tome.
Mr. Joy Bartolome Duldulao of the Philippine Rice Research Institute (PhilRice) and Dr. Lourdes Taylo of the UPLB Institute of Plant Breeding also tackled Malusog Rice and Bt eggplant, respectively. Dr. Clarisse Gonzalvo from the University of the Philippines Los Baños (UPLB) College of Development Communication (Devcom) shared the findings of her survey on biotech farmers' perception of the 2015 biotech ban
“If the precautionary principle tells us ‘when in doubt, stop,' then social science tells us ‘when in doubt, listen'—listen to the farmers whose lives depend on these decisions,” said Dr. Gonzalvo.
A panel discussion with experts from the Bureau of Plant Industry, the Department of Agriculture Biotechnology Program Office, and PhilRice delved into policies and programs that may help catalyze biotech adoption. Dr. Winifredo Dagli, Chair of the UPLB Devcom's Department of Science Communication, moderated the discussion.
For more information, send an email to knowledgecenter@isaaa.org
See https://www.isaaa.org/kc/cropbiotechupdate/article/default.asp?ID=21552
SCIENTIFIC NEWS
Comparative analysis of pattern-triggered and effector-triggered immunity gene expression in susceptible and tolerant cassava genotypes following begomovirus infection
Bulelani L Sizani, Keelan Krinsky, Oboikanyo A Mokoka, Marie E C Rey
Bulelani L Sizani, Keelan Krinsky, Oboikanyo A Mokoka, Marie E C Rey
PLoS One; 2025 Jun 4; 20(6):e0318442. doi: 10.1371/journal.pone.0318442.
.png)
Abstract
South African cassava mosaic virus (SACMV) is one of several bipartite begomoviruses that cause cassava mosaic disease (CMD) which reduces the production yield of the cassava (Manihot esculenta Crantz) crop in many tropical and subtropical regions. SACMV DNA-A and DNA-B encoded-proteins act as virulence factors that aid in inducing different disease severity depending on the host response. Recent evidence suggests a mutual potentiation of cell membrane receptor-associated pattern-triggered immunity (PTI) and nucleotide leucine-rich repeat (NLR) effector-associated immunity (ETI) in plant immune responses. This study aimed to compare expression of SACMV virulence factors, and PTI/ETI, in SACMV-infected susceptible T200 and tolerant TME3 cultivars. Expression of SACMV virulence factors differed between SACMV-infected T200 and TME3 plants at 12, 32 and 67 days post infection (dpi). Notably, at the early stage of infection (12 dpi), expression in TME3 of AV1 and AC2 virulence factors were 10-fold and 30-fold down-regulated, respectively, compared to susceptible T200. At systemic infection (32 dpi) AV1 expression was also significantly lower (4-fold) in TME3 compared to T200. Expression of AC2 (that targets host innate immunity), while significantly lower in both T200 and TME3 at 32 dpi compared to 12 dpi, was also significantly down-regulated (16-fold) in TME3 compared to T200. TME3 recovers around 67 dpi and virus load decreases by 33%, while in T200, symptoms and high SACMV replication persist. Identification and comparison of induced PTI and ETI associated genes upon SACMV-infection in susceptible T200 and tolerant/recovery TME3 cassava genotypes was achieved by whole transcriptome sequencing (RNA-seq) and by reverse transcriptase quantitative PCR (RT-qPCR). Analyses revealed reduced expression of PTI-associated signalling and response genes during SACMV systemic/symptomatic infection (32 dpi) in cassava genotypes. In addition, hydrogen peroxide (H2O2) production, a PTI indicator, was significantly reduced in the symptomatic viral infection stage at 32 dpi. Concurrently at 32 dpi, transcription of ETI signalling and response genes as well as SA biosynthesis and response genes, were upregulated during SACMV systemic infection in TME3. These results indicate that SACMV targets PTI-associated genes during systemic infection at 32 dpi to subvert PTI-mediated antiviral immunity in cassava, which results in reduced induction of ROS production. Differential expression of specific NLR-associated genes also differed between susceptible and tolerant cultivars at 12, 32 and 67 dpi. SACMV virulence factors were shown to play a role in symptom severity in T200 and TME3.
See https://pubmed.ncbi.nlm.nih.gov/40465802/
South African cassava mosaic virus (SACMV) is one of several bipartite begomoviruses that cause cassava mosaic disease (CMD) which reduces the production yield of the cassava (Manihot esculenta Crantz) crop in many tropical and subtropical regions. SACMV DNA-A and DNA-B encoded-proteins act as virulence factors that aid in inducing different disease severity depending on the host response. Recent evidence suggests a mutual potentiation of cell membrane receptor-associated pattern-triggered immunity (PTI) and nucleotide leucine-rich repeat (NLR) effector-associated immunity (ETI) in plant immune responses. This study aimed to compare expression of SACMV virulence factors, and PTI/ETI, in SACMV-infected susceptible T200 and tolerant TME3 cultivars. Expression of SACMV virulence factors differed between SACMV-infected T200 and TME3 plants at 12, 32 and 67 days post infection (dpi). Notably, at the early stage of infection (12 dpi), expression in TME3 of AV1 and AC2 virulence factors were 10-fold and 30-fold down-regulated, respectively, compared to susceptible T200. At systemic infection (32 dpi) AV1 expression was also significantly lower (4-fold) in TME3 compared to T200. Expression of AC2 (that targets host innate immunity), while significantly lower in both T200 and TME3 at 32 dpi compared to 12 dpi, was also significantly down-regulated (16-fold) in TME3 compared to T200. TME3 recovers around 67 dpi and virus load decreases by 33%, while in T200, symptoms and high SACMV replication persist. Identification and comparison of induced PTI and ETI associated genes upon SACMV-infection in susceptible T200 and tolerant/recovery TME3 cassava genotypes was achieved by whole transcriptome sequencing (RNA-seq) and by reverse transcriptase quantitative PCR (RT-qPCR). Analyses revealed reduced expression of PTI-associated signalling and response genes during SACMV systemic/symptomatic infection (32 dpi) in cassava genotypes. In addition, hydrogen peroxide (H2O2) production, a PTI indicator, was significantly reduced in the symptomatic viral infection stage at 32 dpi. Concurrently at 32 dpi, transcription of ETI signalling and response genes as well as SA biosynthesis and response genes, were upregulated during SACMV systemic infection in TME3. These results indicate that SACMV targets PTI-associated genes during systemic infection at 32 dpi to subvert PTI-mediated antiviral immunity in cassava, which results in reduced induction of ROS production. Differential expression of specific NLR-associated genes also differed between susceptible and tolerant cultivars at 12, 32 and 67 dpi. SACMV virulence factors were shown to play a role in symptom severity in T200 and TME3.
See https://pubmed.ncbi.nlm.nih.gov/40465802/
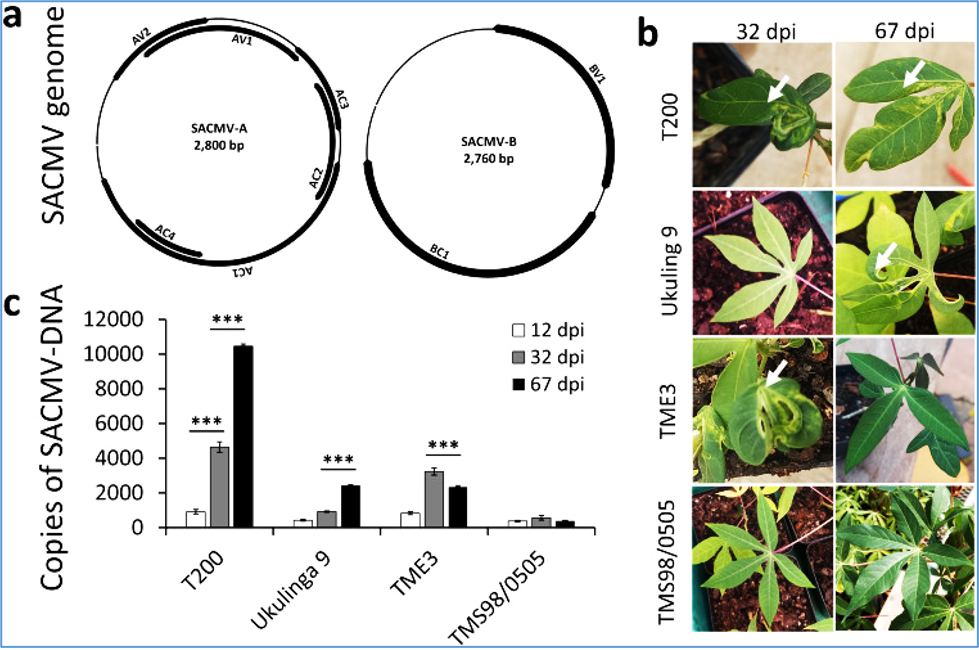
Figure:
South African cassava mosaic virus (SACMV) genome structure and the effect of SACMV on cassava leaf symptoms.
(a) A bipartite ssDNA-A and ssDNA-B genomic structure of SACMV. AC1: replication-associated protein (Rep); AC2: transcriptional activator protein (TrAP); AC3: replication enhancer protein (REn); AC4: silencing suppressor protein); AV1: coat protein (CP); AV2: pre-coat protein; BC1: movement protein (MP) and BV1: nuclear shuttle protein (NSP). (b) Leaf symptoms induced by SACMV manifested by leaf curl and yellow mosaic (arrows). In susceptible T200 symptoms persist at 32 and 67 dpi, while in Ukulinga 9 symptoms appear after 32 dpi (delayed susceptible phenotype). TME3 symptoms manifested at 32 dpi (tolerant, recovery phenotype) but the plant recovered at 67 dpi. TMS98/0505 exhibits no symptoms at any given time point post-infection (resistant genotype). (c) Viral load in T200, Ukulinga 9, TME3 and TMS98/0505 at 12 (pre-symptomatic stage), 32 (symptomatic stage) and 67 (recovery stage) dpi determined as DNA copies of CP using qPCR. Data represent the mean of three independent biological replicates. Error bars represent SD. An asterisk indicates a statistically significant difference according to unpaired Student’s t-test (two-tailed), * p ≤ 0.05. GTPb was used as a housekeeping gene.
.png)











