BULLETIN 1
Mapping for Resilience: How Spatial Data is Transforming Karamoja Cluster
Mapping for Resilience: How Spatial Data is Transforming Karamoja Cluster

CGIAR July 2 2025
Pastoral communities in the Karamoja Cluster (a region spanning Kenya, Uganda, South Sudan, and Ethiopia) are experiencing increased tensions and conflicts over water, pasture and grazing lands
Open data can power smarter decisions. Recognizing the critical role of open data in fostering informed decision-making, That’s why the Humanitarian OpenStreetMap Team (HOTOSM) ESA Hub, through the Beyond the Map Fund, awarded a grant to the International Center for Tropical Agriculture (CIAT). This initiative aimed to leverage Geographic Information Systems (GIS) and remote sensing techniques to map critical pastoral resources such as buildings, roads, towns/markets, waterways and water points across the Karamoja region.
This blogpost explores how this mapping initiative provided critical data to promote resilience and peace building efforts in the region.
Mapping Strategy: Desk-Based Analysis Meets Community Mapping
CIAT and HOTOSM used a complementary two-pronged strategy.
CIAT conducted a comprehensive desk-based spatial analysis of the entire Karamoja Cluster. This included classifying land cover using a Random Forest machine learning model, as well as analysing high-resolution aerial images to map key pastoral resources. Together, these efforts provided a foundational understanding of how pastoral resources are distributed across the region.
In parallel, HOTOSM launched a volunteer mapping campaign focusing on 3 Turkana sub-counties: Turkana West, Turkana Central, and Loima. The campaign adopted a community-centred methodology, and began with the use of the Map Swipe platform to identify building footprints. Subsequent detailed mapping tasks were coordinated through the HOTOSM Tasking Manager, enabling volunteers to contribute localized insights and digitize additional pastoral resources.
This joint approach enhanced the quality and accessibility of the data, while also empowering local stakeholders in the mapping process. The resulting geospatial data lays a vital groundwork for strategic planning, resource allocation, and conflict mitigation—supporting long-term climate adaptation and resilience for the pastoralists of the Karamoja Cluster.
See https://www.cgiar.org/news-events/news/mapping-for-resilience-how-spatial-data-is-transforming-karamoja-cluster/
BULLETIN 2
With Science We Can documentary campaign
With Science We Can documentary campaign

CGIAR July 2 2025
CGIAR has partnered with global knowledge-driven media company, WebsEdge, to produce the With Science We Can Campaign – a multiplatform documentary campaign that will premiere at COP30 in Belém, Brazil.
The CGIAR With Science We Can Campaign will serve as a unique platform to explore how we can achieve a food-secure future through scientific innovation.
The campaign will consist of the flagship 25-minute documentary and the Thought Leadership Film Series. The flagship documentary will take viewers inside CGIAR’s global research centres to reveal science-powered solutions addressing regional agricultural challenges and shaping policy.
To amplify those breakthroughs, a companion Thought-Leadership Film Series will spotlight external innovators— start-ups, policy think-tanks, and grassroots cooperatives—who are translating research into real-world impact.
If you’re interested in getting involved in this campaign, email laila@websedge.com for more information.
See https://www.cgiar.org/news-events/news/with-science-we-can-documentary-campaign/
SCIENTIFIC NEWS
Genome-scale knockout simulation and clustering analysis of drug-resistant breast cancer cells reveal drug sensitization targets
JinA Lim, Hae Deok Jung, Soo Young Park, Moonhyeon Jeon, Da Sol Kim, Ryeongeun Cho, Dohyun Han, Han Suk Ryu, Yoosik Kim, and Hyun Uk Kim
PNAS; June 25, 2025; 122 (26) e2425384122; https://doi.org/10.1073/pnas.2425384122
PNAS; June 25, 2025; 122 (26) e2425384122; https://doi.org/10.1073/pnas.2425384122
Significance
Metabolic reprogramming is a notable phenotype during the acquisition of resistance to chemotherapy drugs. Thus, reverting metabolism can be an effective strategy to sensitize drug-resistant cells. The computational framework developed in this study addresses this challenge by using genome-scale metabolic models and multiomics data. We run single-gene knockout simulations and perform subsequent clustering analysis (UMAP and k -means clustering) to identify drug-sensitizing metabolic gene targets. We demonstrate this approach by taking doxorubicin- and paclitaxel-resistant breast cancer cells as model systems. We find that inhibiting the selected targets in the drug-resistant cells together with doxorubicin or paclitaxel synergistically reduces cell viability. This computational framework allows a cost-efficient identification of effective drug sensitization targets.
Abstract
Anticancer chemotherapy is an essential part of cancer treatment, but the emergence of resistance remains a major hurdle. Metabolic reprogramming is a notable phenotype associated with the acquisition of drug resistance. Here, we develop a computational framework that predicts metabolic gene targets capable of reverting the metabolic state of drug-resistant cells to that of drug-sensitive parental cells, thereby sensitizing the resistant cells. The computational framework performs single-gene knockout simulation of genome-scale metabolic models that predicts genome-wide metabolic flux distribution in drug-resistant cells, and clusters the resulting knockout flux data using uniform manifold approximation and projection, followed by k-means clustering. From the clustering analysis, knockout genes that lead to the flux data near that of drug-sensitive cells are considered drug sensitization targets. This computational approach is demonstrated using doxorubicin- and paclitaxel-resistant MCF7 breast cancer cells. Drug sensitization targets are further refined based on proteome and metabolome data, which generate GOT1 for doxorubicin-resistant MCF7, GPI for paclitaxel-resistant MCF7, and SLC1A5 as a common target. These targets are experimentally validated where treating drug-resistant cancer cells with small-molecule inhibitors results in increased sensitivity of drug-resistant cells to doxorubicin or paclitaxel. The applicability of the developed framework is further demonstrated using drug-resistant triple-negative breast cancer cells. Taken together, the computational framework predicts drug sensitization targets in an intuitive and cost-efficient manner and can be applied to overcome drug-resistant cells associated with various cancers and other metabolic diseases.
See https://www.pnas.org/doi/10.1073/pnas.2425384122
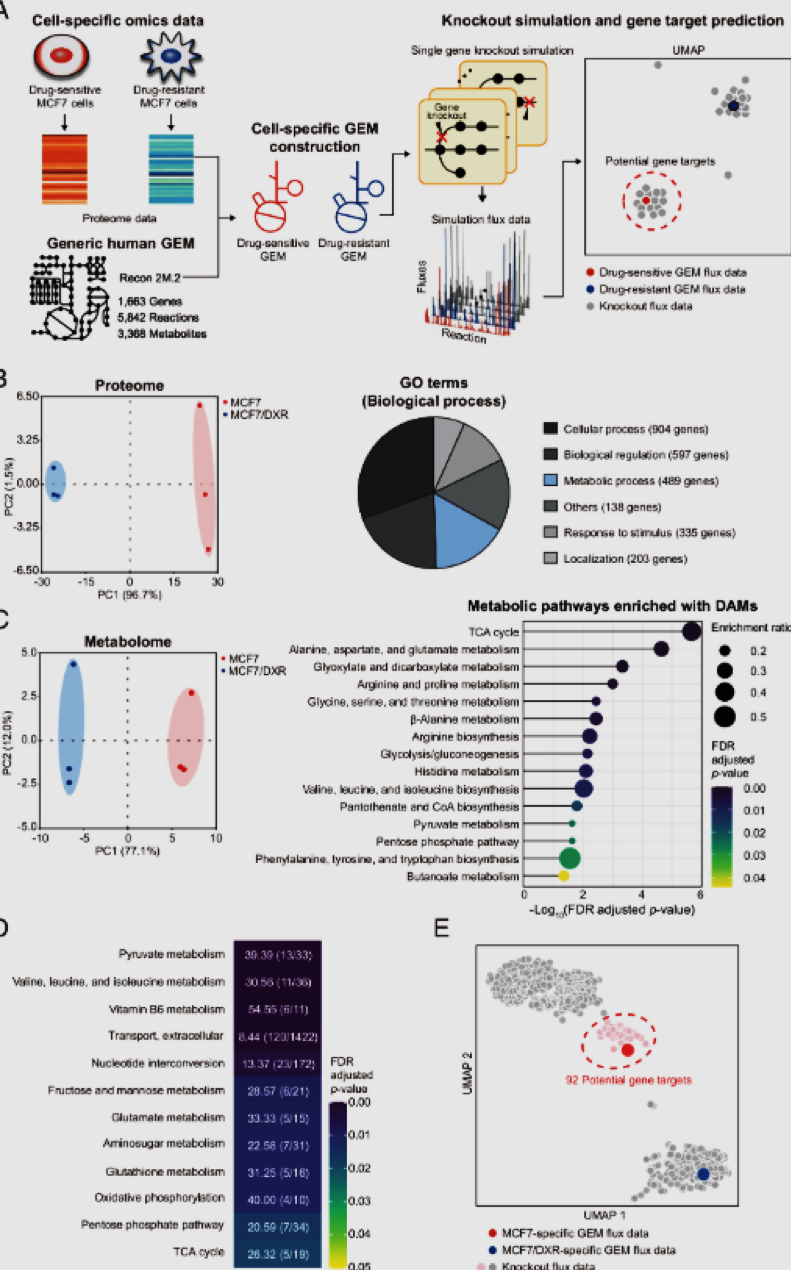
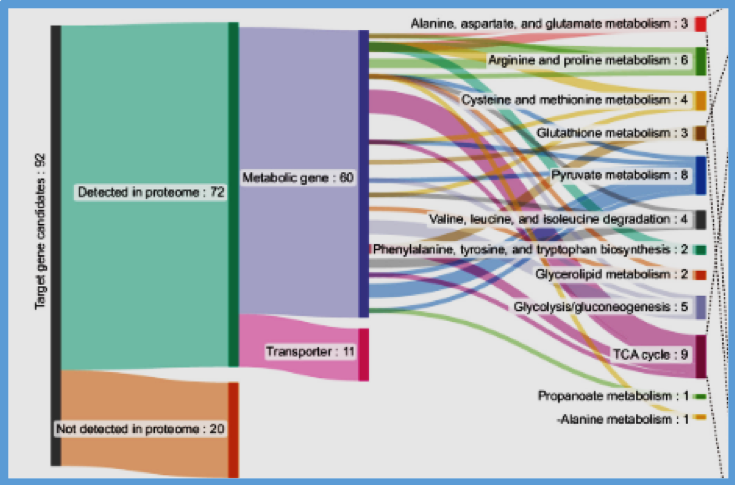
FIGURE:
The overall scheme of predicting drug-sensitizing gene targets for MCF7/DXR. (A) Workflow of reconstructing and simulating cell-specific GEMs to predict gene targets that can sensitize the MCF7/DXR. Proteome data previously obtained for MCF7 and MCF7/DXR (28) were used to build cell-specific GEMs. The MCF7/DXR-specific GEMs were subjected to single-gene knockout simulation. The resulting knockout flux data were subjected to UMAP and k-means clustering. Knockout genes that clustered with that of MCF7 (red in the UMAP plot) were considered drug sensitization targets. (B) PCA (Left) and GO enrichment analysis (Right) of proteome data from MCF7 and MCF7/DXR. Enriched GO terms on biological processes of significant DEPs are shown on the Right. (C) PCA (Left) and KEGG enrichment analysis (Right) of metabolome data from MCF7 and MCF7/DXR. KEGG pathways significantly enriched with differentially abundant metabolites (DAMs) are shown on the Right. (D) Metabolic pathways significantly enriched with different reaction fluxes between MCF7 and MCF7/DXR, identified through flux enrichment analysis using their specific GEMs. The numbers indicate the ratio of reactions with different fluxes to the total number of reactions in each pathway. (E) UMAP plot of the knockout flux data as well as those from the MCF7- and MCF7/DXR-specific GEMs. Knockout genes corresponding to the knockout flux data clustered with the MCF7 flux data were considered potential drug sensitization targets (dotted red ellipse).
The overall scheme of predicting drug-sensitizing gene targets for MCF7/DXR. (A) Workflow of reconstructing and simulating cell-specific GEMs to predict gene targets that can sensitize the MCF7/DXR. Proteome data previously obtained for MCF7 and MCF7/DXR (28) were used to build cell-specific GEMs. The MCF7/DXR-specific GEMs were subjected to single-gene knockout simulation. The resulting knockout flux data were subjected to UMAP and k-means clustering. Knockout genes that clustered with that of MCF7 (red in the UMAP plot) were considered drug sensitization targets. (B) PCA (Left) and GO enrichment analysis (Right) of proteome data from MCF7 and MCF7/DXR. Enriched GO terms on biological processes of significant DEPs are shown on the Right. (C) PCA (Left) and KEGG enrichment analysis (Right) of metabolome data from MCF7 and MCF7/DXR. KEGG pathways significantly enriched with differentially abundant metabolites (DAMs) are shown on the Right. (D) Metabolic pathways significantly enriched with different reaction fluxes between MCF7 and MCF7/DXR, identified through flux enrichment analysis using their specific GEMs. The numbers indicate the ratio of reactions with different fluxes to the total number of reactions in each pathway. (E) UMAP plot of the knockout flux data as well as those from the MCF7- and MCF7/DXR-specific GEMs. Knockout genes corresponding to the knockout flux data clustered with the MCF7 flux data were considered potential drug sensitization targets (dotted red ellipse).
.png)










