BULLETIN 1
NATURE+ in Vietnam: Country report 2022-2024
NATURE+ in Vietnam: Country report 2022-2024

Figure: Northern Vietnam landscapes where NATURE+ scientists characterized the agrobiodiversity and farmers’ plots.
CGIAR Feb 20 2025
CGIAR’s Nature-Positive Solutions Initiative made substantial progress in Northern Vietnam, alongside rural, Indigenous communities who were the Initiative’s main partners. The hilly landscapes are rich in threatened and understudied agrobiodiversity, which some communities still successfully cultivate. To help landholders address soil degradation and unsustainable agricultural intensification, NATURE+ and partners studied traditional crops, promoted their sustainable use and conservation through value chains, and supported circular bioeconomy practices. Additional work focused on true-cost accounting for food systems and landscape restoration. The Initiative’s work in Vietnam is confidently positioned for continuity and growth as part of the CGIAR Research Program 2025-2030.
https://www.cgiar.org/news-events/news/nature-in-vietnam-country-report-2022-2024/
https://www.cgiar.org/news-events/news/nature-in-vietnam-country-report-2022-2024/
BULLETIN 2
NATURE+ in India: Country Report 2022-2024
NATURE+ in India: Country Report 2022-2024
CGIAR; Feb 20 2025

The CGIAR Nature-Positive Solutions had several successes in India. Most of the Initiative’s key activities happened in tribal areas in Maharashtra State. As with much of NATURE+’s work –
particularly on neglected and underutilized species, or NUS – traditional knowledge was key to the Initiative’s activities on crops, trees and nature-positive agriculture in India. The Initiative’s highlights included launching a national hub for circular bioeconomy, supporting traditional seed banks, and integrated watershed management systems. This report covers NATURE+ activities in India, which set the stage for continuity in the CGIAR Research Portfolio 2025-2030.
See https://www.cgiar.org/news-events/news/nature-in-india-country-report-2022-2024/
particularly on neglected and underutilized species, or NUS – traditional knowledge was key to the Initiative’s activities on crops, trees and nature-positive agriculture in India. The Initiative’s highlights included launching a national hub for circular bioeconomy, supporting traditional seed banks, and integrated watershed management systems. This report covers NATURE+ activities in India, which set the stage for continuity in the CGIAR Research Portfolio 2025-2030.
See https://www.cgiar.org/news-events/news/nature-in-india-country-report-2022-2024/
SCIENTIFIC NEWS
DArTseq-based silicoDArT and SNP markers reveal the genetic diversity and population structure of Kenyan cashew (Anacardium occidentale L.) landraces
Dennis Wamalabe Mukhebi, Pauline Wambui Gachanja, Diana Jepkoech Karan, Brenda Muthoni Kamau, Pauline Wangeci King'ori, Bicko Steve Juma, Wilton Mwema Mbinda
PLoS One; 2025 Jan 31; 20(1):e0313850. doi: 10.1371/journal.pone.0313850.
Dennis Wamalabe Mukhebi, Pauline Wambui Gachanja, Diana Jepkoech Karan, Brenda Muthoni Kamau, Pauline Wangeci King'ori, Bicko Steve Juma, Wilton Mwema Mbinda
PLoS One; 2025 Jan 31; 20(1):e0313850. doi: 10.1371/journal.pone.0313850.

Abstract
Cashew (Anacardium occidentale L.) is an important tree grown worldwide for its edible fruits, nuts and other products of industrial applications. The ecologically sensitive cashew-growing region in coastal Kenya is significantly affected by rising temperatures, droughts, floods, and shifting rainfall patterns. These changes adversely impact cashew growth by altering flowering patterns, increasing pests and diseases, and causing postharvest losses, which ultimately result in reduced yields and tree mortality. This is exacerbated by the long juvenile phase, high heterozygosity, lack of trait correlations, large mature plant size, and inadequate genomic resources. For the first time, the Diversity Array Technology (DArT) technology was employed to identify DArT (silicoDArT) and single nucleotide polymorphisms (SNPs) markers for genomic understanding of cashew in Kenya. Cashew leaf samples were collected in Kwale, Kilifi and Lamu counties along coastal Kenya followed by DNA extraction. The reduced libraries were sequenced using Hiseq 2500 Illumina sequencer, and the SNPs called using DarTsoft14. A total of 27,495 silicoDArT and 17,008 SNP markers were reported, of which 1340 silicoDArT and 824 SNP markers were used for analyses after screening, with > 80% call rate, > 95% reproducibility, polymorphism information content (PIC ≥ 0.25) and one ratio (>0.25). The silicoDArT and SNP markers had mean PIC values ranging from 0.02-0.50 and 0.0-0.5, with an allelic richness ranging from 1.992 to 1.994 for silicoDArT and 1.862 to 1.889 for SNP markers. The observed heterozygosity and expected values ranged from 0.50-0.55 and 0.34-0.37, and 0.56-0.57 and 0.33 for both silicoDArT and SNP markers respectively. Understanding cashew genomics through the application of SilicoDArT and SNP markers is crucial for advancing cashew genomic breeding programs aimed at improving yield and nut quality, and enhancing resistance or tolerance to biotic and abiotic stresses. Our study presents an overview of the genetic diversity of cashew landraces in Kenya and demonstrates that DArT systems are a reliable tool for advancing genomic research in cashew breeding.
See https://pubmed.ncbi.nlm.nih.gov/39888943/
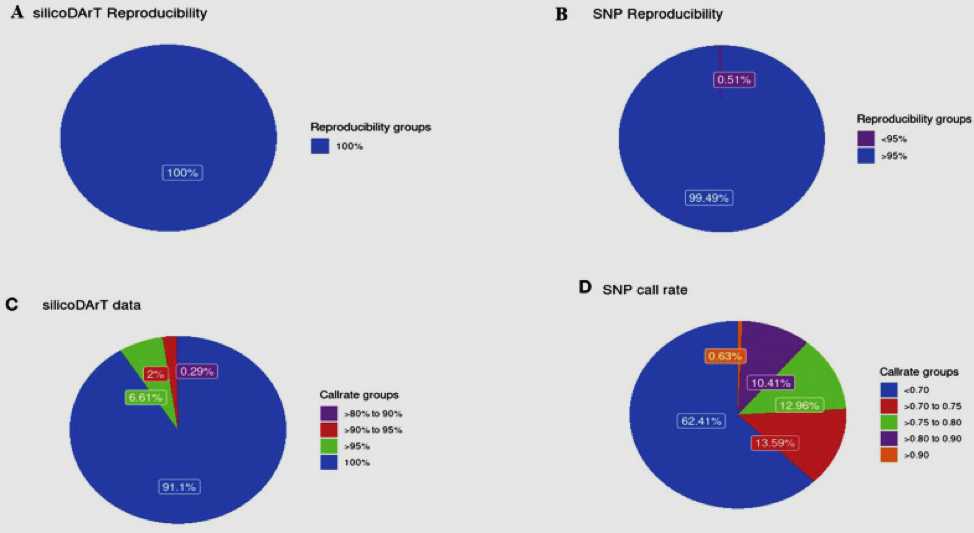
Figure: Distribution of marker data for reproducibility and call rate.
A. Reproducibility of SilicoDArT markers, B. Reproducibility of SNP markers. C. Call rates of SilicoDArT markers, D. Call rates of SNP markers.
.png)












